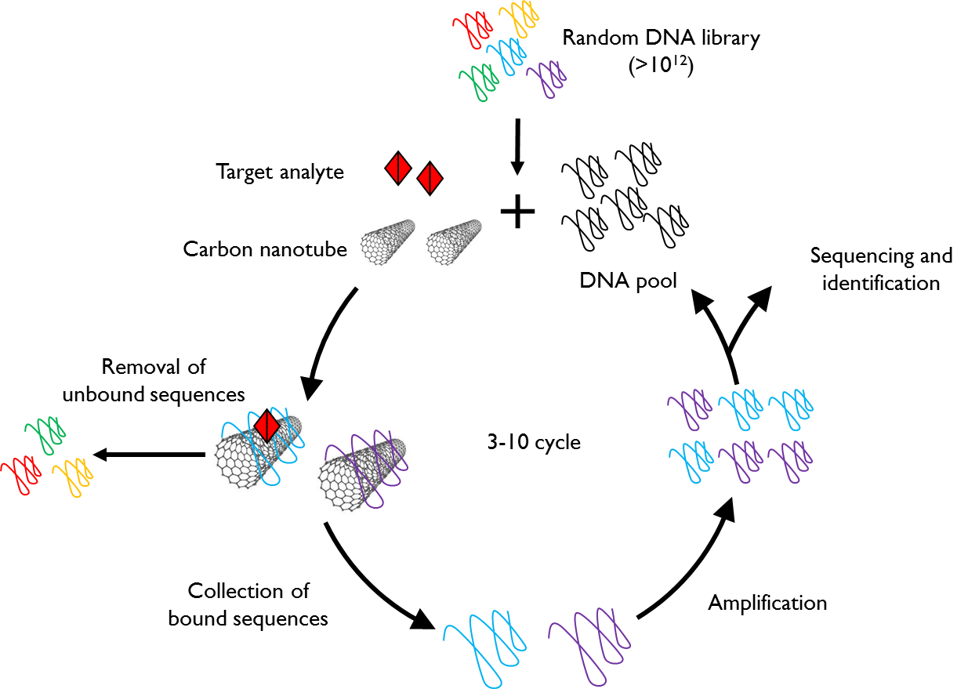
High-Throughput Evolution of Nanosensors
Chemically-specific imaging of small molecules in complex biological media is often accomplished with synthetic nanosensors. In most embodiments of nanosensor platforms, the principal challenge lies in the laborious process of identifying synthetically-produced selective and sensitive molecular recognition elements. We’ve devised an evolution-based platform, in which 1012 unique nanotube-pinned polynucleotide polymers can be screened for their ability to bind a desired analyte when adsorbed to carbon nanotubes, and provide a selective near-infrared fluorescence signal in its presence. With a combination of polymerase chain reaction, and next-generation sequencing, we can identify nanotube-polynucleotide conjugates selective and sensitive for analytes such as neuromodulator serotonin. We are exploring this approach to expand the library of synthetic nanosensors to image neurotransmitters, neuromodulators, and neuropeptides.
Using chemometric approaches, including principal component analysis and hierarchical clustering, we can analyze fluorescence spectra obtained from initial library screens to identify new and unique analyte-nanosensor hits. Additionally, this analysis can inform the relationship between chemical properties of the analyte and the fluorescence response of the nanosensors. To date, we have used this approach to characterize a near-infrared nanosensor for the chemotherapeutic doxorubicin.
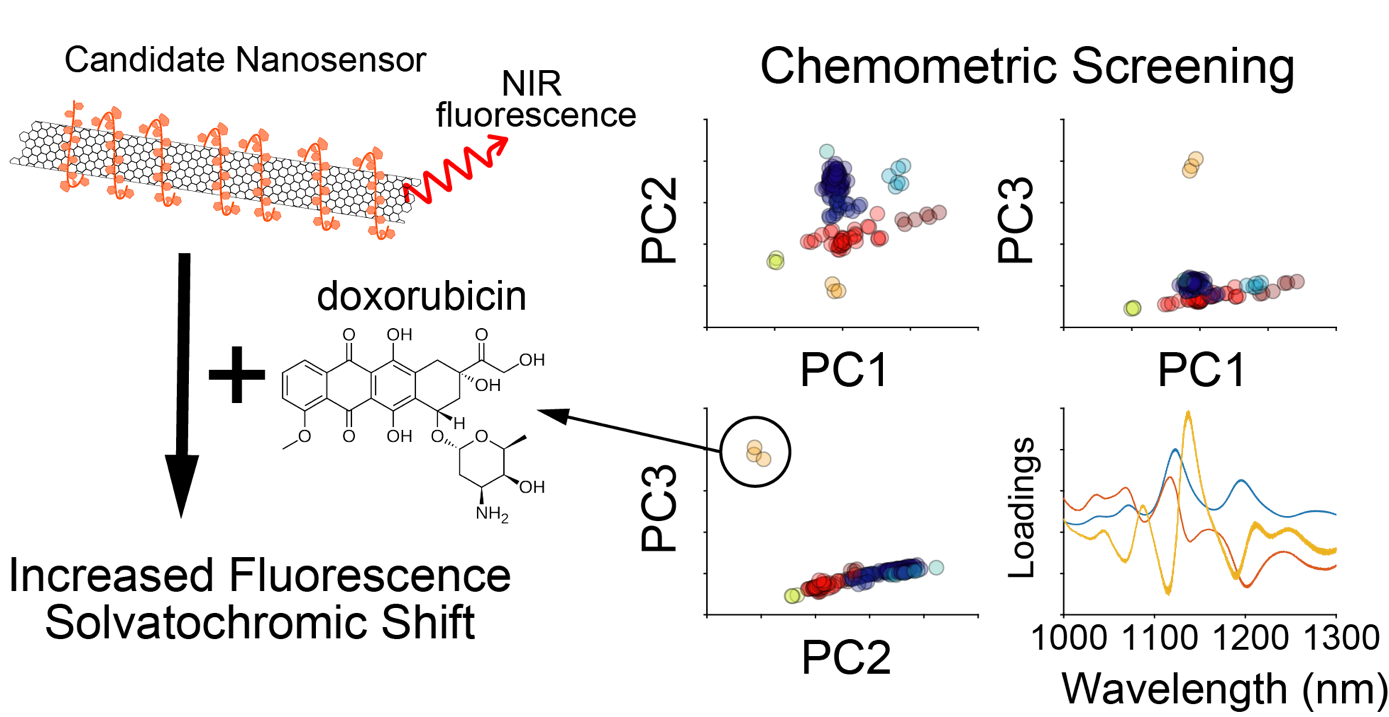
.
Understanding Sensor Dynamics
Engineered nanomaterials are promising investigational tools for biological sensing and imaging applications due to their distinctive optical and physical characteristics. The critical – and often overlooked – challenge with such novel tools is bridging the gap between their in vitro synthesis and their in vivo implementation. To this end, our lab seeks to unearth the fundamental mechanisms of interaction between nanoparticle sensors, and the systems they are designed to probe. When a nanoparticle enters a biological fluid, the surface becomes coated with proteins to form the “corona”.
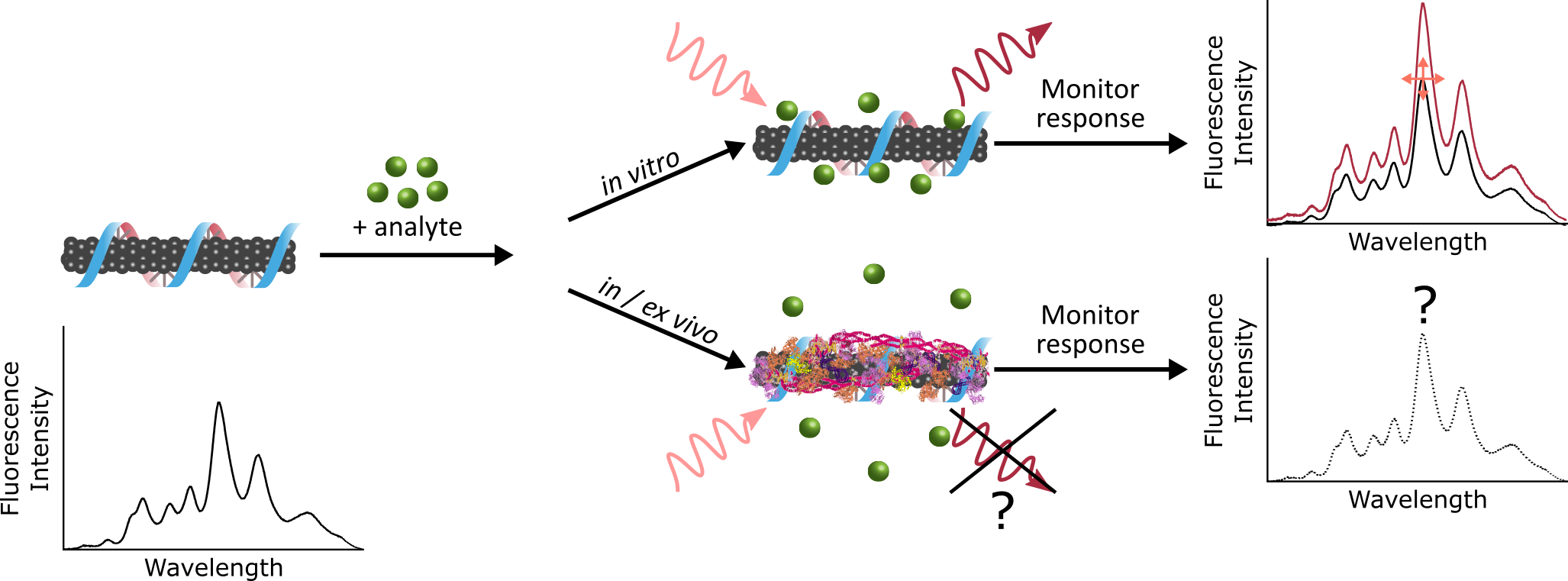
Binding of proteins to the nanoparticle leads to significant consequences, including masking of the as-designed sensing functionality, and direction nanoparticle fate. As we move towards applying nanotechnology in vivo, we aim to build a framework for elucidating how our engineered nanoparticles are affecting, and being affected by, complex biological environments. This entails characterizing the composition, thermodynamics, and kinetics of nanoparticle-protein corona formation to gain a holistic understanding of the system that will, in turn, be applied to inform future nanotechnology design strategies.
.
Nature Mimetics for Protein Sensing
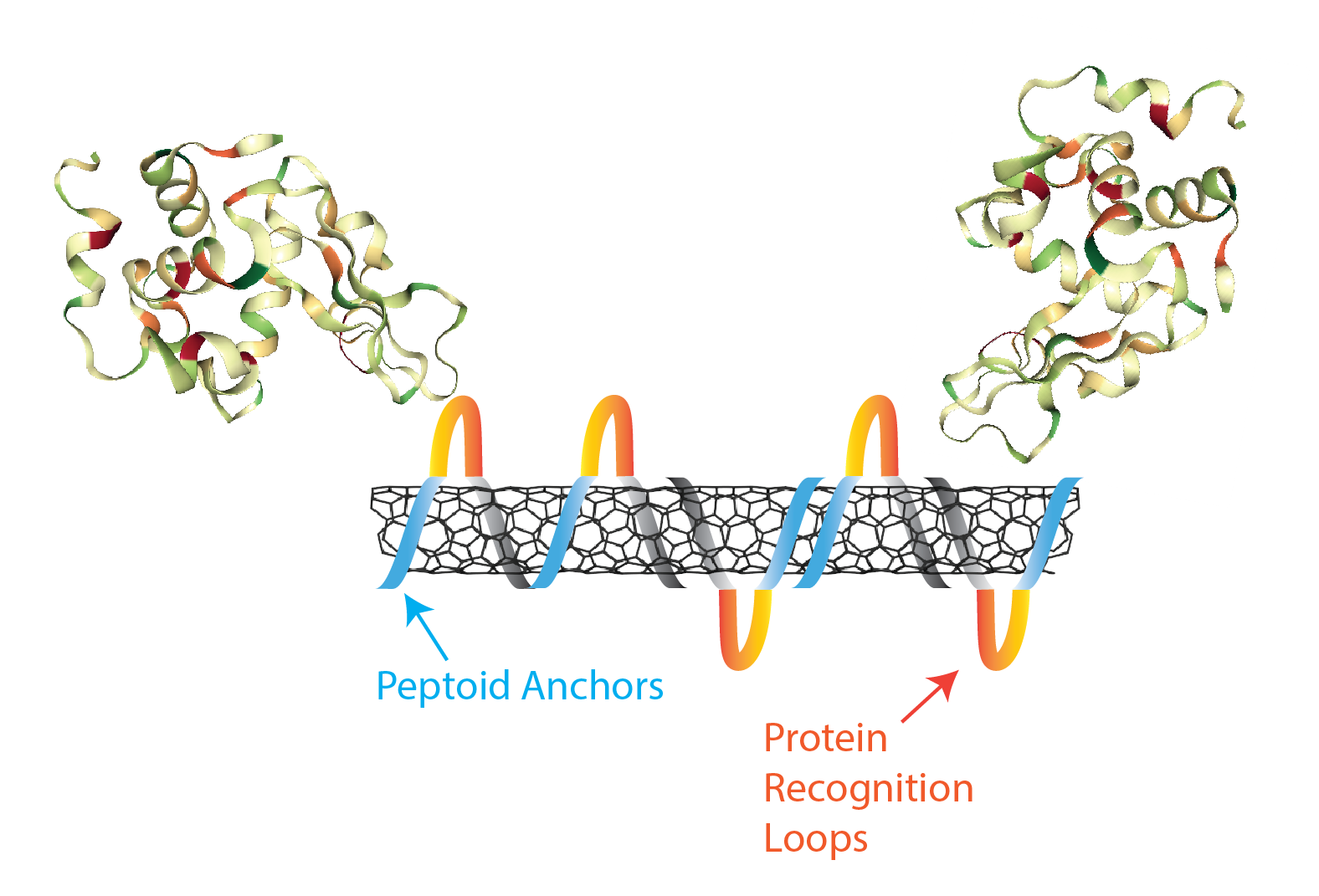
Nanomaterials are of a similar size to the molecular entities that govern life within cells, and are therefore well-suited to studying life at the sub-cellular scale. To this end, our laboratory couples biomimetic polymers with single-walled nanotubes for the creation of protein optical sensors, a type of ‘synthetic antibody’. We are developing antibody-mimetic binding loops with peptoid, N-substituted glycine polymers, that exhibit a much larger monomer space than peptides, and are more stable than their peptide counterparts.
.
Enzyme-Immobilization for Biological Sensing

We develop preparation methods for enzyme-immobilized semiconducting single-walled carbon nanotubes. Amphiphilic block copolymers are used to both 1) wrap with the surface of carbon nanotubes and 2) capture enzymes by noncovalent interaction with enzyme surface functional groups. By redox reaction of various types of enzymes with their target single molecules, NIR-II fluorescence of enzyme-SWNT nanosensors changes with excellent selectivity. These enzyme-SWNT nanosensors could be further used as in vivo biosensors and bioimaging probes.
Scientists: Jaquesta Adams, Francis Ledesma, Alison Lui, Jaewan Mun, Nicole Navarro, Shoichi Nishitani, Moein Safaee, Dabin Yim.
Select Publications: Del Bonis et al. ACS Biochemistry (2018) || Beyene et al. Nano Letters (2018) || Chio et al. Nano Letters 2019 || Jeong et al. Science Advances 2019 || Beyene et al. Science Advances (2019) || Pinals et al. JACS 2020 || Chio et al. Advanced Functional Materials (2020) || Pinals et al. Angewandte Chemie (2020) || Pinals ACS Nano (2021)|| Ouassil et al. Science Advances (2022)
